Taxonomy
Description
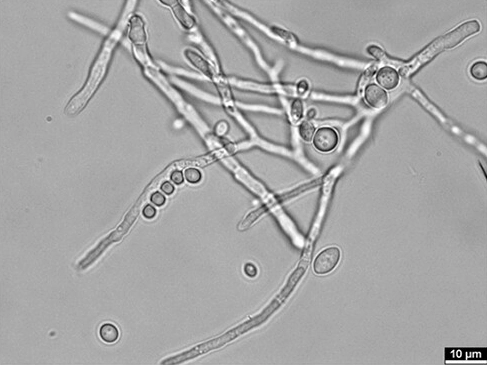
Blastobotrys mokoenaii belongs to a category of species that are referred to within yeast biotechnology as non-conventional yeasts (Mukherjee et al., 2017). Here, non-conventional implies that they might have other phenotypes compared to the industrial workhorse Saccharomyces cerevisiae, such as improved tolerance to process conditions, or different metabolic activities. Yeasts from the Blastobotrys genus are of interest for biotechnological applications due their oleaginous (lipid-accumulating) nature and their broad substrate range (Sanya et al., 2021). B. mokoenaii NRRL Y-27120 is for instance capable of rapid growth on the polysaccharide xylan (Ravn et al., 2023).
The B. mokoenaii NRRL Y-27120 genome assembly that is displayed here on the Genome Portal was generated in the Y1000+ Project (Opulente et al. 2024, Shen et al., 2018). The annotation of protein-coding genes was generated in Sweden in a study by the Geijer research group at Chalmers University of Technology.
How to cite
If you use the genome assembly presented in the genome portal from this species in your research, please cite the original publication:
Shen, X.-X., Opulente, D. A., Kominek, J., Zhou, X., Steenwyk, J. L., Buh, K. V., Haase, M. A. B., Wisecaver, J. H., Wang, M., Doering, D. T., Boudouris, J. T., Schneider, R. M., Langdon, Q. K., Ohkuma, M., Endoh, R., Takashima, M., Manabe, R., Čadež, N., Libkind, D., … Rokas, A. (2018). Tempo and Mode of Genome Evolution in the Budding Yeast Subphylum. Cell, 175(6), 1533-1545.e20. https://doi.org/10.1016/j.cell.2018.10.023If you use the annotation of protein-coding genes, which was generated by a different research group, please cite:
Ravn, J.; Sörensen Ristinmaa, A.; Mazurkewich, ScotS.t; Borges Dias, G.; Larsbrink, J.; Geijer, C. (2025). Gene annotation of Blastobotrys mokoenaii, Blastobotrys illinoisensis, and Blastobotrys malaysiensis. Chalmers University of Technology. Dataset. https://doi.org/10.17044/scilifelab.28606814.v1If you have used the pages for this species in the Genome Portal, please refer to it in-text as: “The Blastobotrys mokoenaii entry in the Swedish Reference Genome Portal (Retrieved ).” and use the following for the bibliography:
Swedish Reference Genome Portal (Retrieved ), SciLifeLab Data Centre, version 1.7.0 from https://genomes.scilifelab.se, RRID:SCR_026008References
Mukherjee, V., Radecka, D., Aerts, G., Verstrepen, K. J., Lievens, B., & Thevelein, J. M. (2017). Phenotypic landscape of non-conventional yeast species for different stress tolerance traits desirable in bioethanol fermentation. Biotechnology for Biofuels, 10(1), 216. https://doi.org/10.1186/s13068-017-0899-5
Opulente, D. A., LaBella, A. L., Harrison, M.-C., Wolters, J. F., Liu, C., Li, Y., Kominek, J., Steenwyk, J. L., Stoneman, H. R., VanDenAvond, J., Miller, C. R., Langdon, Q. K., Silva, M., Gonçalves, C., Ubbelohde, E. J., Li, Y., Buh, K. V., Jarzyna, M., Haase, M. A. B., … Hittinger, C. T. (2024). Genomic factors shape carbon and nitrogen metabolic niche breadth across Saccharomycotina yeasts. Science, 384(6694), eadj4503. https://doi.org/10.1126/science.adj4503
Ravn, J. L., Sörensen Ristinmaa, A., Coleman, T., Larsbrink, J., & Geijer, C. (2023). Yeasts Have Evolved Divergent Enzyme Strategies To Deconstruct and Metabolize Xylan. Microbiology Spectrum, 11(3), e00245-23. https://doi.org/10.1128/spectrum.00245-23
Ravn, J.; Sörensen Ristinmaa, A.; Mazurkewich, ScotS.t; Borges Dias, G.; Larsbrink, J.; Geijer, C. (2025). Gene annotation of Blastobotrys mokoenaii, Blastobotrys illinoisensis, and Blastobotrys malaysiensis. Chalmers University of Technology. Dataset. https://doi.org/10.17044/scilifelab.28606814.v1
Sanya, D. R. A., Onésime, D., Passoth, V., Maiti, M. K., Chattopadhyay, A., & Khot, M. B. (2021). Yeasts of the Blastobotrys genus are promising platform for lipid-based fuels and oleochemicals production. Applied Microbiology and Biotechnology, 105(12), 4879–4897. https://doi.org/10.1007/s00253-021-11354-3
Shen, X.-X., Opulente, D. A., Kominek, J., Zhou, X., Steenwyk, J. L., Buh, K. V., Haase, M. A. B., Wisecaver, J. H., Wang, M., Doering, D. T., Boudouris, J. T., Schneider, R. M., Langdon, Q. K., Ohkuma, M., Endoh, R., Takashima, M., Manabe, R., Čadež, N., Libkind, D., … Rokas, A. (2018). Tempo and Mode of Genome Evolution in the Budding Yeast Subphylum. Cell, 175(6), 1533-1545.e20. https://doi.org/10.1016/j.cell.2018.10.023
Changelog
- 10/06/2025 - Update track name from Protein-coding genes to Genes to reflect that the annotation also contains ncRNA genes
- 05/05/2025 - Species first published on the Portal
Page last updated: 05/05/2025